Paper Summary: U-Net
U-nets yielded better image segmentation in medical imaging. U-Net: Convolutional Networks for Biomedical Image Segmentation paper was published in 2015.
Problem
There is large consent that successful training of deep networks requires many thousand annotated training samples. The paper presents a network and training strategy that relies on the strong use of data augmentation to use the available annotated samples more efficiently.
The typical use of convolutional networks is on classification tasks, where the output to an image is a single class label. However, in many visual tasks, especially in biomedical image processing, the desired output should include localization i.e. a class label is supposed to be assigned to each pixel. Moreover, thousands of training images are usually beyond reach in biomedical tasks.
Solution
The U-Net architecture is built upon the Fully Convolutional Network and modified in a way that it yields better segmentation in medical imaging. The paper use’s excessive data augmentation by applying elastic deformations to the available training images. This allows the network to learn invariance to such deformations, without the need to see these transformations in the annotated image corpus.
Architecture
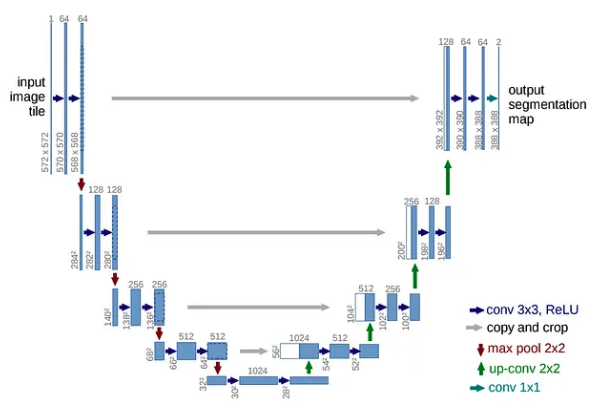 The u-net comprises of two parts an encoder/contraction path(left side) and a decoder/expansion path(right side).
The u-net comprises of two parts an encoder/contraction path(left side) and a decoder/expansion path(right side).
Contraction path consists of a repeated application of a 3x3 convolutions(unpadded) each followed by a ReLU and a 2x2 max pooling operation with stride 2 for downsampling. At each downsampling step, we double the number of feature channels. This captures context via a compact feature map.
The expansion path consists of upsampling of the feature map followed by a 2x2 convolution(“up-convolution”) that halves the number of feature channels a concatenation with the cropped feature map from the contracting path, and a 3x3 convolutions, followed by a ReLU. The upsampling of the feature dimension is done to meet the same size as the block to be concatenated on the left.
The expansion increases the “what” which helps in getting more features but losses the localization, localization information is concatenated from the contraction path.
The cropping is necessary due to the loss of border pixels in every convolution. At the final layer, a 1x1 convolution is used to map each 64-components feature vector to the desired number of classes. In this case, it is 2 as the output feature map has 2 classes; cells and membrane.
The main contribution of this paper
a. Overlap-tile strategy
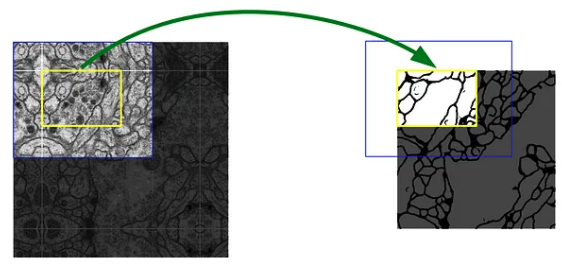
Prediction of the segmentation in the yellow area requires image data within the blue area as input. missing input data is extrapolated by mirroring, this is used to predict pixels in the border region of the image.
b. Data augmentation by applying elastic deformations to training images.
This allows the network to learn invariance to such deformations, without the need to see these transformations in the annotated image corpus. This is important in biomedical segmentation since deformation is the most common variation in tissue and realistic deformations can be simulated efficiently.
c. Separation of touching objects of the same class.
This is done using a weighted loss, where the separating background labels between touching cells obtain a large weight in the loss function. This force the network to learn the small separation borders between touching cells.
Real world application of this contribution
Thousands of training images are beyond reach in biomedical tasks and require experts and take lots of time to annotate. This could automate the process thus lowering the cost and time it takes to annotate.
This can also be applied in other areas such as quality control and inspection and manufacturing.
In this paper, the techniques were applied in the segmentation of neuronal structures in electron microscopic recordings, cell segmentation task in light microscopic images and HeLa cells on a flat glass recorded by differential interference contrast microscopy.
References
[1] Ronneberger, O., Fischer, P., & Brox, T. (2015, October). U-net: Convolutional networks for biomedical image segmentation.
Back to learning!